Structure Waves in Biological Systems: Evidence for deBroglie Waves in Protein Crystals
Click here to view a message from Dr. Gerald Pollack, Editor-in-Chief
John Grant Watterson
18 Tomanbil Terrace, Ashmore, 4214, Australia
(retired: formerly, Department of Science Griffith University, Southport, 4217, Australia)
email: jgwatterson@gmail.com
Keywords: water clusters, structure wave, proteins, α helix, β sheet
Submitted: January 31, 2024
Revised: January 5, 2025
Accepted: February 10, 2025
Published: March 12, 2025
1. Abstract
The cluster model of liquid structure explains the origin of the forces that are responsible for osmotic phenomena. In contrast to the many textbook explanations based on thermal motion of solute molecules, the model, based on coherent co-operative motion of solvent molecules (water), is shown to explain also other hydration mechanisms, including protein function. Its central concept, the pressure pixel, is defined as the volume occupied by a molecule in a perfect gas (about 40 cubic nm at ambient conditions), which is assumed to be the smallest volume in which pressure is exerted in liquids also. The collision mechanism of the familiar kinetic theory of gases is replaced by co-ordinated motion of molecules of solution propagating as a structure wave. It is shown how, when clusters of different solutions in contact carry and exchange equal energies (kT), cluster size, as judged by wavelength, is predicted to be in the 1 nm size range and obey the deBroglie relation. In addition, it is emphasized that long-standing results from X-ray, electron and neutron scattering give the fundamental protein domain size also in this range. This physical property indicates that both water clusters and protein domains, support the same wave motion. It is further concluded that oscillation of the H-bonded internal structures of domains must therefore also be periodic, with α-helices performing longitudinal and β-sheets performing transverse motion in synch with the structure wave of the whole medium.
2. Introduction
The oft quoted phrase that water is essential for life is normally interpreted to mean that water is the universal medium in which life originated and evolved. However, in this presentation, water is not viewed as the medium, but as the agent. On this watery planet osmotic mechanisms show their presence in the creation of vast deposits of clay soils, in the regulation of turgor in plants, and in the ability of cells to control the dynamics of the gelled state of their cytoplasm. On the other hand, osmotic mechanisms can also be a nuisance to us when, for example, grass roots crack the garden path, or swelling soils cause buildings to move, and we must manage the high pressures needed to run filtration in desalination plants.
The most popular explanation (among many) found in physics texts, asserts that osmosis is “a diffusion process.” It is the result of collisions caused by thermal molecular motion in liquids (water) – that is, diffusion – which produces flow against pressure and compensates for entropy increases. However, both these textbook explanations contradict bedrock principles of physics. The first (diffusion against pressure) contravenes Newton’s second law of motion, and the second (build-up of solution pressure) contravenes the thermodynamic law of minimization of free energy. So, to proceed we first need to know the answer to the question: What then does cause pressure in liquids?
Pressure is a macroscopic phenomenon. As Pascal taught, pressure is transmitted from wall to wall throughout a fluid body – or put another way, each wall of its container feels the presence of the others. The Kinetic Theory of Gases explains pressure in terms of molecular collisions on the micro level; however, this interpretation cannot be extended to liquids, where the close-packed molecules are bonded to one another by tensile forces. In the cluster-wave model, it is collision of the structure wave with the boundaries, as opposed to speeding gas molecules, that causes pressure in liquids.
This hypothesis means that it is the wave rather than molecules, that carries the impulses and to comply with Pascals Principle, the wave must travel back and forth in all directions throughout the medium and be reflected at its boundaries. In contrast to gases then, the mechanism at the micro level in liquids resembles the way force is produced by a spring pressing against the walls of its container, whereby each wavelength can be pictured as one turn of the spring. We imagine that when the spring is squeezed, liquid media come under increased pressure – or the more turns in the spring, the smaller their size, but the greater their force. According to this model, the space occupied by one turn and the energy it carries defines the pressure pixel. Clusters are mesoscopic entities above the level of molecules, which are both particles and waves occupying a 3D shape in the medium. They are the physical manifestation of the pressure pixel, whose size is equivalent to the volume occupied by a single gas molecule in an ideal gas, u = kT/P (Watterson, 1987).
It follows that the concept of a cluster used here differs from the commonly accepted meaning, which depicts a group of molecules bonded together acting as a unit particle. In the present model on the other hand, the meaning refers to a regular geometric arrangement with quasi crystalline, as opposed to random, structural properties. Hence a moving, or flickering, cluster is to be visualized as a moving pattern, not a moving collection of molecules. In fact, in osmotic processes, such geometric solvent clusters can translate to-and-fro in either direction between solution media in contact to achieve equilibrium (from low to high pressure, or vice versa) without flow of solvent mass. Moving patterns constitute a wave, and this wave provides the basis for the transfer of momentum and energy.
In 1987, I proposed that osmotic equilibrium between two solutions (commonly separated by a semipermeable membrane) can be explained using concepts analogous to those we use as the basis of the kinetic theory of gases. Equilibrium occurs under those conditions where the frequency, f, of clusters crossing through the membrane from each side, and the quantum of energy, kT, carried by each cluster are equal (Watterson, 1987).
f = n1v1 = -n2v2 = v1/u1 = -v2 /u2 Equation 1
and kT = P1u1 = P2u2 = M1v1 = M2v2 Equation 2
where n, v, u, M, and P represent concentration, velocity, volume, momentum, and pressure of clusters carried by the structure wave in the direction normal to the membrane boundary. These two conditions mean that the net energy flux, fkT, across the boundary is zero, since v2 and M2 are opposite in sign (negative) to v1 and M1. Calling on the gas analogy again here, these equations can be interpreted in terms of velocities of two different types of gas molecules, with that of the more concentrated one traveling at the slower speed to give the same striking rate on the boundary. Likewise, those molecules with higher speed will have lower momentum to keep the kinetic energy of the two gases equal. But this analogous picture is purely imaginary. Molecules of different gases cannot cross the boundary and mix into each other, because they cannot change their velocities or momenta. A more reasonable analogy depicts the gas molecules colliding at the boundary surface and rebounding off it rather than crossing through it and mixing. In this case, the dynamic parameters are described by the familiar textbook example of particle collision.
3. The Gas Analogy
When two elastic particles, masses m1 and m2, collide, they rebound according to the laws of conservation of energy and momentum. We begin using the ideal model of two billiard balls, which is the favored scenario of physics texts illustrated in Figure 1. Their average velocity is given by
v = (m1v1 + m2v2)/(m1 + m2) Equation 3
where v1 and v2, are their velocities and the difference in their velocities is given by
d = (v1 – v2)/(m1 + m2) Equation 4
normalized to total mass (m1 + m2).
Straightforward algebra shows that their combined kinetic energies is given by
KE = m1v12/2 + m2v22/2 Equation 5
= (m1 + m2)v2/2 + (m1 + m2) m1m2d2/2 Equation 6
= E + D Equation 7
This result describes the ideal case of a collision of perfectly elastic balls. In real (non-elastic) events, the kinetic energy is not conserved, and KE after the collision is given by E, plus a variable amount of D, of which some or all is lost as heat. In the favored example you find in physics textbooks, velocities are measured by the observer relative to the fixed billiard tabletop, assumed to be stationary. This means that, since E depends on the frame of reference, the energies represented by E are relative energies, while those represented by D are absolute, since they depend on the difference (v1-v2) only, which has a constant value whether the billiard table is moving or not.
The reference frame condition of interest to us is when balls collide with equal and opposite momenta, since then their average velocity is zero
m1v1 = -m2v2 Equation 8 v = 0 Equation 9
and the sum of their kinetic energies, m1v12/2 and m2v22/2, equals D since now E=0. However, at the instant of the collision the energy of each flows into the opposite ball resulting in a uniform energy density throughout the combined single mass (m1+m2)
m2v22/2m1 = m1v12/2m2
= (m1v12 + m2v22)/2(m1+m2)
= D/(m1+m2) Equation 10
from the algebraic identity, a/b = c/d = (a+c)/(b+d).
This result indicates that at this instant, every molecule of the material of the balls has the same energy, whether originating in m1 or m2. And importantly, these algebraic terms no longer represent kinetic energies, but rather they represent a summed elastic wave resulting from transduction of mechanical kinetic energies down to micro molecular vibrations. In the case of perfect elastic collisions, rebound then follows when the wave is reflected off the balls’ internal surfaces, as it splits into the two separate kinetic energies again. Therefore, the analogy tells us that, during this event, the original kinetic energies were both transduced (changed hierarchical level) and transferred (swapped between balls). We will see later that this result introduces the concept of a transduction mechanism applicable to protein function.
To examine this scenario a little further, we imagine the case of inelastic balls that fuse on impact. Here, the resulting fused mass, (m1+m2), stops moving as before, and the total kinetic energy KE, again equal to D, is transferred down into frictional molecular motion. In such non-elastic collisions, molecular vibrations are non-coherent and do not sum to produce a macro elastic wave that causes the rebound. We normally expect such non-coherence to produce heat, i.e., infrared radiation, but of course the wavelength of emitted radiation depends on the strength of the impact. This result describes a dispersion step, in which macro kinetic energy is dissipated down into the micro level, and then even to the nano level of radiation.
The molecules in both balls all arrive at the point of impact at the same instant, because they constitute mathematically ideal masses of zero volume rather than extended 3D volumes occupied by clusters in a liquid medium. Volumes must take time (period) to pass through the boundary depending on the shape of the wave (wavelength). The close parallel of the gas analogy to osmotic mechanisms becomes clear, when we expand the point masses (billiard balls) m1 and m2 in space, so that they now each occupy unit volume as in a perfect gas. In this expanded model, the variables n1 and n2 of Equation 1 now represent concentrations of single isolated molecules colliding with the boundary separating two gases
f m0 = (n1m0)v1 = -(n2m0)v2 Equation 11
= m1v1 = -m2v2 Equation 8
where m0 is the molecular mass of the material constituting the balls, so that Equation 8 now refers to unit volume. This identity shows us that the interpretation in terms of concentrations of gas molecules is equivalent to that of solid balls with equal but opposite momenta, even though the collision event is now imagined as being diluted in time and space – an instant becomes period and a point becomes wavelength. See Figure 2 for a simplified model of energy concentrations distributed within a spatial grid.

In summary:
To compare the three analagous events, we began by describing energy transfer between two solid bodies (billiard balls), called particle collision (Fig. 1) governed by Equation 8. In this case the exchange between the two particles happens instantaneously on impact and results in the molecules of both particles sharing equal energy given by Equation 10.
Next, this result is the same as that describing the exchange between two gases of different concentrations and pressures (Fig. 3). The molecules of both gases have the same (average) energy, kT, which they exchange one-for-one with frequency given by Equations 1 and 11.
Finally, an identical spatial grid of two solutions as that of two gases is used to model osmotic equilibrium assuming that the volume occupied by each gas molecule is the same as the volume of the pressure pixel in a liquid occupied by the cluster wave (Fig. 4).
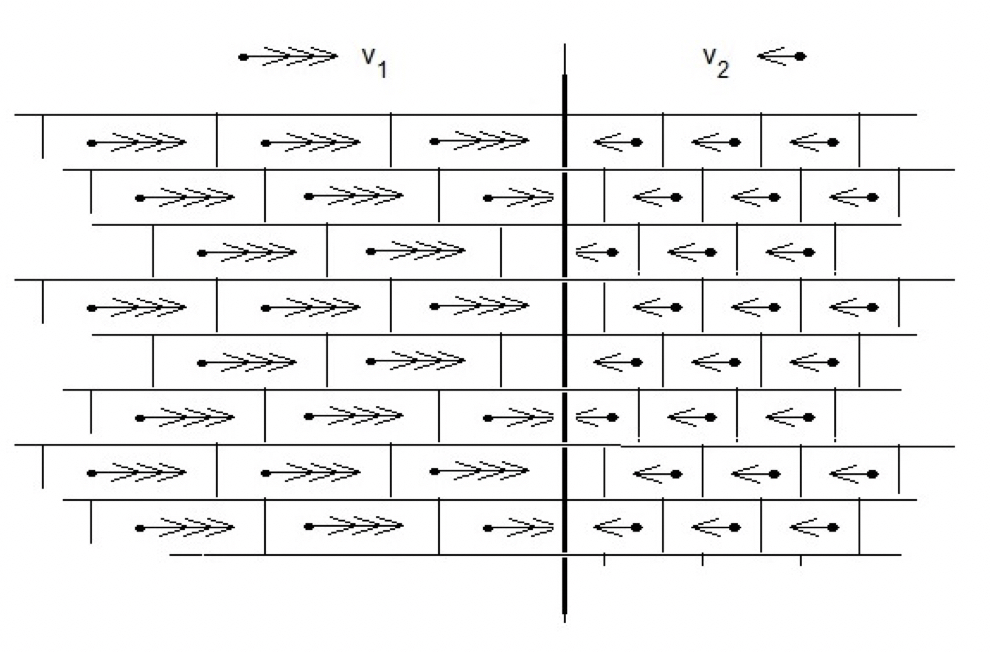
4. Osmosis
At the liquid boundary, the structure waves mutually share their energies as did the colliding mass particles (billiard balls) as shown in Equation 10. However, motion does not reverse at the boundary causing the waves to rebound off one another but continues on into the opposite medium. Unlike the case of colliding particles that must retain their masses following the rebound event, here they are not entities with mass, but the oscillating geometric structural patterns described above in the Introduction. Thus, clusters leaving one liquid medium can adjust their dynamic parameters to satisfy conditions in the new medium they enter.
Here, there is no momentum in the familiar sense of the mechanical concept, M = mv, because the molecules of the solutions are not in macro translational mode. Liquid may, or may not, flow across the boundary, but when equilibrium is reached there is no longer net translation of mass. As noted above, n1 and n2, now refer to cluster concentrations, i.e., structural entities with volumes, not mass entities. They are rather pixels of volume, u = a λ, where a is the cross-section area of the wave’s forward movement across the boundary, and λ its wavelength, and the momentum that each carries is now smoothed out as a density rather than existing in a point. Since they flow into each other mutually at a rate of one-for-one, their cross-sections are equal in area and we have from the frequency in Equation 1
v1 / λ1 = v2 / λ2 Equation 12
and hence from Equation 2
M1 λ1 = M2 λ Equation 13
Showing that cluster size as measured by λ is determined by momentum according to the deBroglie relation.
This analysis attributes a vector property to cluster dynamics. In fact, the vectorial direction was already assumed in the discussion on the force exerted at the boundary, that is, normal to the membrane. On the other hand, deBroglie’s hypothesis of 1924 defined a momentum wave from his analysis of quantum levels in atomic orbitals, and thus the present derivation is independent of his argument. Therefore, by analogy, the cluster model leads to the conclusion that liquid structure may also be a quantum phenomenon (Fig. 4).
In the traditional osmotic experiment, the solvent phase (usually water) denoted below as solution 1, is separated from solution 2 (say, salt in water) by a semi-permeable membrane. Solution 1 is open to the atmosphere and therefore atmospheric pressure, whereas solution 2 is confined. Osmotic equilibrium is reached after solvent flows through the membrane into solution 2 raising the pressure therein resulting in the inequality P 2 > P 1 , giving the familiar relation for osmotic pressure
Π = P 2 – P 1 Equation 14
Then from Equation 2 Π = (n 2 – n 1) kT Equation 15
showing that osmotic pressure is the result of an increase in cluster concentration. Comparison with van’t Hoff’s familiar equation identifies (n2-n1) as the increase in solute molecule concentration in solution 2 above solution 1. I’ve shown elsewhere that, since n and u are the inverse of one another, the increase corresponds to decreasing wavelength, and hence an increase in the number of nodes (or antinodes) in the wave. Furthermore, I argued how this decrease in cluster size is the result of the disruptive effect that foreign molecules must have on structure in the solvent medium. And we now see how this change is accompanied by an increase in cluster momentum in the solution, leading in turn to the measured increase in pressure there (Watterson, 1995).
The traditional formula quoted in Equation 14 is a special case because P1 is always taken to be the pressure on pure solvent under laboratory conditions. A more general statement describing the phenomenon is the relation between two solutions which are in contact but do not mix,
P2 / P1 = n2 / n1 = λ1 / λ2 Equation 16
At normal laboratory conditions, P=1 atm and T=300 K,
u = kT/P = a λ = 4 10-26 = 40 nm3. Equation 17
This value is, of course, an average volume since we expect the structure in pure liquid medium (solvent/water) to be distributed over a wide range of sizes. In bulk medium under general isotropic conditions in the absence of solute surfaces, the average cubic volume has an edge of the order of 3 nm.
Spectroscopic studies are widely used in the search for the answer to whether liquids are structured or not. The controversial co-existence of two distinct densities in water (LDW and HDW) at ambient conditions, and the search for the associated thermodynamic parameter – their critical point (LLCP) – is now more than two decades old! (Huang et al., 2009; Voeikov and delGiudice, 2009; Jansson et al., 2010; Nilsson et al., 2012; Musumeci et al., 2012; Taschin et al., 2012; Soper, 2013). Extensive neutron and electron scattering studies have so far failed to yield a clear answer; indeed, many researchers have concluded that spectroscopic studies can be interpreted in terms of a homogeneous medium without postulating the existence of clustering. Neutron diffraction is a particularly direct method as it can be applied at normal laboratory conditions (Towey et al., 2016; Teixeira, 2017). The energy of the radiation at ambient temperature (room kT) corresponds to a wavelength in the 1 nm range. On the other hand, electrons need to be accelerated to an energy about 50 times higher to achieve a comparable wavelength, being some 2000 times lighter than neutrons. Recent reviews discuss many of the proposals in support of either the one state or two state model. However, whichever interpretation is eventually agreed to, it is clear that this technique reveals the existence of a collective species of water molecules in this size range. But, as explained in the Introduction, the model under discussion in this presentation is not in line with the picture of two distinct density species because it takes a wave form. As originally proposed in 1981, this picture likens an aqueous medium to a continuous spring with the dynamic properties that sustain the propagation of an energy wave and its associated structural fluctuations rather than independent islands of different densities.
Further strong evidence is given by the existence of the hydration force, which shows that ordered layers with thickness in the nm range form at hydrophilic surfaces (Israelachvilli, 1992). These structures exhibit properties expected of internally bonded clusters, since they actively exclude solutes from the zone adjacent to the surface. Many researchers have dubbed the water composing these layers “exclusion zone” water. Additionally, the layer thickness can be reversibly controlled and monitored by applying IR radiation – a property demonstrating that clusters are dynamic entities (Chai et al., 2009; Chen et al., 2012; Hwang et al., 2018). The hydration force is thus a non-random force able to do work by exerting lateral tension parallel to the interface and outward pressure normal to the interface – the possibility of complex vector action we will meet again in the next section. Such dynamic properties invite the obvious conclusion that clusters were involved in the origins of prebiotic life (interested readers can find further discussion of this idea on my website, https://www.thewaterpixel.com).
5. Protein Crystallization
An unexpected, even surprising, property of proteins is that the size of their basic building block, a protein domain, has this dimension also. It is the case quite independently of function, even though it contravenes the postulates of statistical thermodynamics. This uniformity of molecular structure shown by a population of such a vast number of isolated chain molecules is the underlying explanation for the appearance of neutron scattering patterns observed in protein solutions decades ago (Giordano et al., 1990), and for the unexpected phenomenon of protein crystallization, which happens at normal laboratory temperatures. In fact, the first X-ray structures obtained by the groups of Kendrew and Perutz were both single domain molecules of dimensions in the 1 nm range (Kendrew et al., 1958; Perutz et al., 1960). The spatial regularity achieved in samples of such vast numbers as they crystallize can be accounted for by the transmission of vectorial information across and throughout the intervening medium – a structure wave. In addition to spatial regularity, such large molecules composed of over 1000 atoms each need controlled coherence as they adopt their orientation to allow crystals to form, which must be achieved by long-range forces present in the medium suppressing the background of disruptive thermal motion. I have discussed this model in extenso elsewhere (Watterson, 2024).
As far as I am aware, there is no accepted explanation of protein crystallization based on the principles of thermodynamics. Direct physical attraction between molecules does not exist – in fact, there is no obvious contact between them, as crystals contain a high-water content, which can be as much as 50% w/w, with the result that their physical state is best described as a soft gel. When we consider the vast number of molecules involved together with the degree of precision that results, protein crystallization is perhaps the most spectacular example of the creation of order in a simple solution induced by molecular structure in water. Such observations give the strong indication that the explanation must involve the reasons for the well-known (and often unwanted by experimentalists) tight water-protein interactions. Those interactions are not attractive forces in the sense we normally think of (e.g., ionic, H-bonds . . .), but a force generated on a higher hierarchical level exemplified by spatial fitting, thereby helping to intensify a geometric grid already present as a nascent form in the pure medium.
Another promising avenue is the comparison of protein crystals with water samples studied under negative pressures (Bergonzi et al., 2016). Since the pressure inside a crystal is negative, or close to zero, information on molecular interactions in water prepared under tension (sometimes called “stretched water”) in the absence of protein, should be applicable to our models of how the spontaneous growth of crystals happens and to explaining their stability once formed.
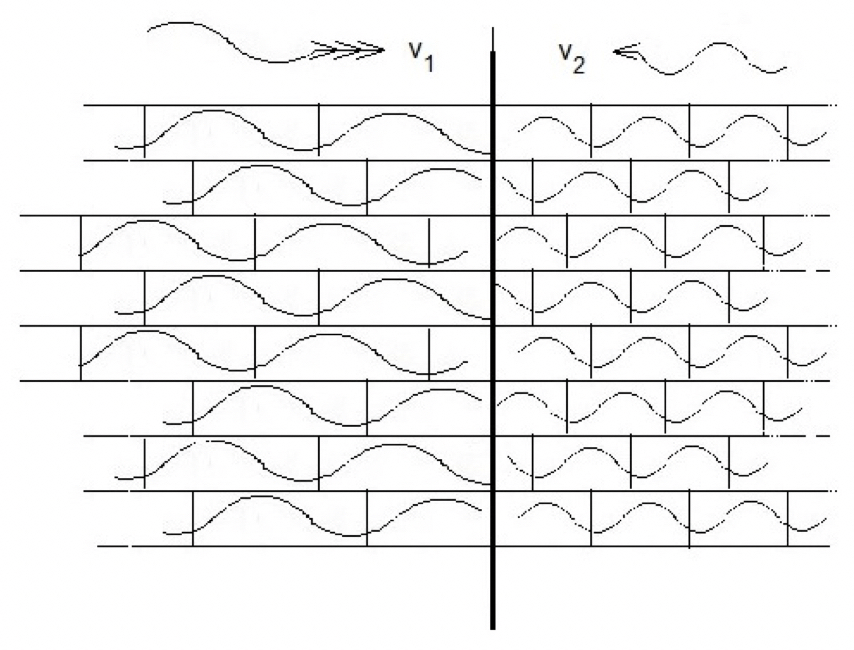
The pattern of H-bonds in a particular protein is covalently fixed, whereas in liquid water it is not. Indeed, the internal structures of over 100,000 individual proteins are today already known [see Protein Data Bank (www.PDB.org)]. On the other hand, the arrangement of bonds in liquid water is flexible, allowing them to re-align according to the pattern set by the presence of a fixed protein domain, and in this way support the passage of the wave. In proteins the arrangement consists of groupings of two simple structural elements, α-helices and β-sheets, each 5-10 H-bonds in length spanning 1-3 nm, of which the H-bonds in helices are positioned in series and those in sheets are in parallel (Fig. 5). Such simple spatial arrangements suggest collective oscillations supporting longitudinal and transverse motion, respectively (Zolotaryuk et al., 1999; Zolotaryuk and Salerno, 2006; Brizhik et al., 2009). Indeed, both types of motion occurring simultaneously (called LO-TO splitting), have been observed in water on the time scale of Debye relaxations (Nibali et al., 2014; Elton and Fernandez-Serra, 2016).
This hypothetical mechanism of mutual influences between protein domains and cluster waves – protein structures inducing cluster wave shape on the one hand, and wave propagation stimulating domain crystallization on the other – suggests the further implication that the origin of the protein domain is to be found in water because of the multitude of variations it offers. Readers fond of music need only remember that the same note sounds different when played on different instruments, and that talented players can even identify overtones responsible for the difference.
Mathematical readers on the other hand, interpret this sonic phenomenon in terms of the detail they find hidden in Fourier analysis, where variations in amplitudes and frequencies lead to almost endless harmonic possibilities. I feel that these modes of thinking are analogous to the way in which protein chemists depict internal bonding as folded ribbons held together by the fixed non-random pattern of the basic α and β structures. So, by extension, a cluster of 1500 water molecules with flexible bonding have the possibility of adopting arrangements that oscillate in tune with that fixed internal protein pattern. Such knowledge would hopefully throw light on the mystery of the uniqueness and stability of the protein fold – perhaps the most basic question in all of biology.
Conclusion
This paper opens by quoting the popular belief that water is the necessary environment for the origin of life and closes with the predication that the internal structure of proteins facilitates the transmission of an energy-carrying structure wave. It is argued that water is not just the happy chaotic medium for the appearance of life, but a medium composed of molecular order underlying the coordinated activity we associate with living matter.
In this model, clusters are not seen as fixed groupings of molecules, but as quasi crystalline geometric arrangements that move through the medium. This movement takes the form of a wave, which is responsible for exerting forces in liquids as opposed to the mechanism of collisions between molecules that we know operates in gases. Clusters are the manifestation of energy quanta defined by the pressure pixel – the smallest volume experiencing the force of pressure – the volume occupied by a molecule in a perfect gas, i.e., about 40 cubic nm (a cube with an edge of 3.3 nm) under normal laboratory conditions. Thus, this picture of a cluster is that of a moving volume rather than a moving mass. The comparison of how pressure is exerted in a liquid with that in a gas is illustrated diagrammatically in Figures 3 and 4.
The gas analogy also provides an example of the crucial dynamic of energy transfer. When two particles of different masses collide, simple algebra shows they share their energies. At the moment of impact, the energy of one flows into the other resulting in a uniform common energy given by Equation 10. In the language of physics, their kinetic energies are transduced into the new form of potential spring energy of the combined mass, before reverting into the kinetic energies as they rebound off one another. In a continuous medium on the other hand, the cluster-wave model holds that, rather than colliding, clusters travel through each other and cross the boundary (semi-permeable membrane) as they move to-and-fro between solutions. At osmotic equilibrium there is no net transfer of energy, so the clusters must cross from either side at the same frequency. This crucial condition expressed in Equation 1 leads directly to the deBroglie relation which determines their wavelengths. Results from osmosis set the sizes of clusters by modeling the medium in a simplified depiction as a grid of cubes in the 1 nm range in agreement with the above calculation based on the gas equation.
This contribution places great emphasis on results from structural studies in the biological field of X-ray crystallography. This technique has established the spatial regularity seen in thousands of protein crystals. The similarity in form and physical dimensions shown by protein crystals to the array of pressure pixels in osmotic systems indicates that the crystalline protein gels are constructed of clusters fitting together with protein domains. Furthermore, the wave mechanism of pressure in liquids, transduction between kinetic and elastic energies, scattering studies and the 3D conformations adopted by protein crystals, taken together suggest that such observations must arise from the same underlying phenomenon. The thread linking them is the transmission of a wave. In liquids, molecules oscillate cooperatively forming coherent clusters that adopt size and shape in response to pressure conditions. On the other hand, in protein molecules the internal bonding is fixed; however, synchronized oscillations of the two basic bonding motifs, α-helical and β-sheets, support the longitudinal and transverse modes of wave motion. Therefore, this mechanism operating in biological systems ensures that the same molecular movement is transmitted through both physical and biological materials.
Ethical Statement
Funding: none
Conflict of Interest: N/A
Ethical approval: N/A
Informed consent: N/A
Author contribution: JGW is the sole author
Data availability: N/A
References
Bergonzi I, Mercury L, Simon P, Jamme F, Shmulovich K (2016). Oversolubility in the microvicinity of solid-solution interfaces. Phys Chem Chem Phys 18: 14974-14885.
Brizhik LS, Eremko AA, Piette BMAG, Zakrzewski WJ (2009). Direct transport of Davydov solitons by unbiased forces, in: Russo, N., (Ed), Self-organization of molecular systems. NATO Science series A: Chemistry and Biology, Springer, pp. 89-101.
Chai B, Yoo H, Pollack GH (2009). Effect of radiant energy on near-surface water. J Phys Chem B 113: 13953-13958.
Chen CS, Chung WJ, Hsu IC, Wu CM, Chin WC (2012). Force field measurements within the exclusion zone of water. J Biol Phys 38: 113-120.
Elton D and Fernandez-Serra M (2016). The hydrogen-bond network of water supports propagating optical phonon-like modes. Nature Comm 7:10193, pp.1-5.
Giordano R, Salvato G, Wanderlingh F, Wanderlingh U (1990). Quasielastic and inelastic neutron scattering in macromolecular solutions. Phys Rev A 41: 689-696.
Huang C, Wikfeldt KT, Tokushima T, Nordlund D, Harada Y, Bergmann U, Niebuhr M, Weiss TM, Horikawa Y, Leetmaa M, Ljungberg MP, Takahashi O, Lenz A, Ojamae L, Lyubartsev AP, Shin S, Pettersson LGM, Nilsson A (2009). The inhomogeneous structure of water at ambient conditions. PNAS USA 106: 15214-15218.
Hwang SG, Hong JK, Sharma A, Pollack GH, Bahng GW (2018). Exclusion zone and heterogeneous water structure at ambient temperature. Plos One 13(4): e0195057.
Israelachvilli J (1992). Intermolecular and surface forces. Academic Press, New York.
Jansson H, Bergman R, Swenson J (2010). Hidden slow dynamics in water. Phys Rev Lett 104(1): 017802
Kendrew ES, Bodo G, Dintizis HM, Parrish RG, Wyckoff H, Phillips DC (1958). A three-dimensional model of the myoglobin molecule obtained by X-ray analysis. Nature 181: 662-666.
Medvedev KE, Kinch LN, Grishin NV (2018). Functional and evolutionary analysis of viral proteins containing a Rossmann-like fold. Protein Sci 27: 1450-1463.
Medvedev KE, Kinch LN, Schaeffer RD, Pei J, Grishin NV (2021). A fifth of the protein world: Rossmann-like proteins as an evolutionarily successful structural unit. J Mol Biol 433: 166788.
Musumeci F, Grasso R, Lanzano L, Scordino A, Triglia A, Tudisco A, Gulino M (2012). Delayed fluorescence: a novel technique to obtain new insights into water structure. J Biol Phys 38: 181-195.
Nibali VC, Dangelo G, Paciaroni A, Tobias DJ, Tarek M (2014). On the coupling between the collective dynamics of proteins and their hydration water. J Phys Chem Lett 5: 1181-1186.
Nilsson A, Huang C, Pettersson L (2012). Fluctuations in ambient water. J Mol Liquids 176: 2-16.
Perutz M, Rossmann MG, Cullis AF, Muirhead H, Wills G, North ACT (1960). Structure of haemoglobin. Nature 185: 416-422.
Soper A K (2013). Radical reappraisal of water structure in hydrophilic confinement. Chem Phys Lett 590: 1-15.
Taschin A, Bartolini P, Eramo R, Righihi R, Torre R (2013). Evidence of two distinct local structures of water from ambient to supercooled conditions. Nature Comm 2401: 1-8.
Teixeira J (2017). The contribution of small angle and quasi-elastic scattering to the physics of liquid water. J Phys Conf Series 848: 01203-01208.
Towey JJ, Soper AK, Dougan L (2016). Low density water structure observed in a nanosegregated cryoprotectant solution at low temperatures from 285 to 238K. J Phys Chem B 120: 4439-4448.
Voeikov VL and del Giudice E (2009). Water respiration – The basis of the living state. Water 1:52-57.
Watterson JG (1987). Solvent cluster size and colligative properties. Phys Chem Liquids 16: 317-320.
Watterson JG (1997). The Pressure Pixel – Unit of Life. BioSystems 41: 141-152.
Watterson JG (1995). What Drives Osmosis? J Biol Phys 21: 1-9.
Watterson JG, (2024). The cluster model of energy transduction in biological systems. https://doi.org/10.1016/j.biosystems.2024.105213
Zolotaryuk AV, Christiansen PL, Norden B, Savin AV (1999). Soliton and ratchet motions in helices. Conden Matter Phys 2: 293-302.
Zolotaryuk Y and Salerno M (2006). Discrete såoliton ratchets driven by biharmonic fields. Phys Rev E 73: 066621
